iTaxoTools
iTaxotools is a bioinformatic platform designed to facilitate the core work of taxonomists, that is, delimiting, diagnosing and describing species.
About
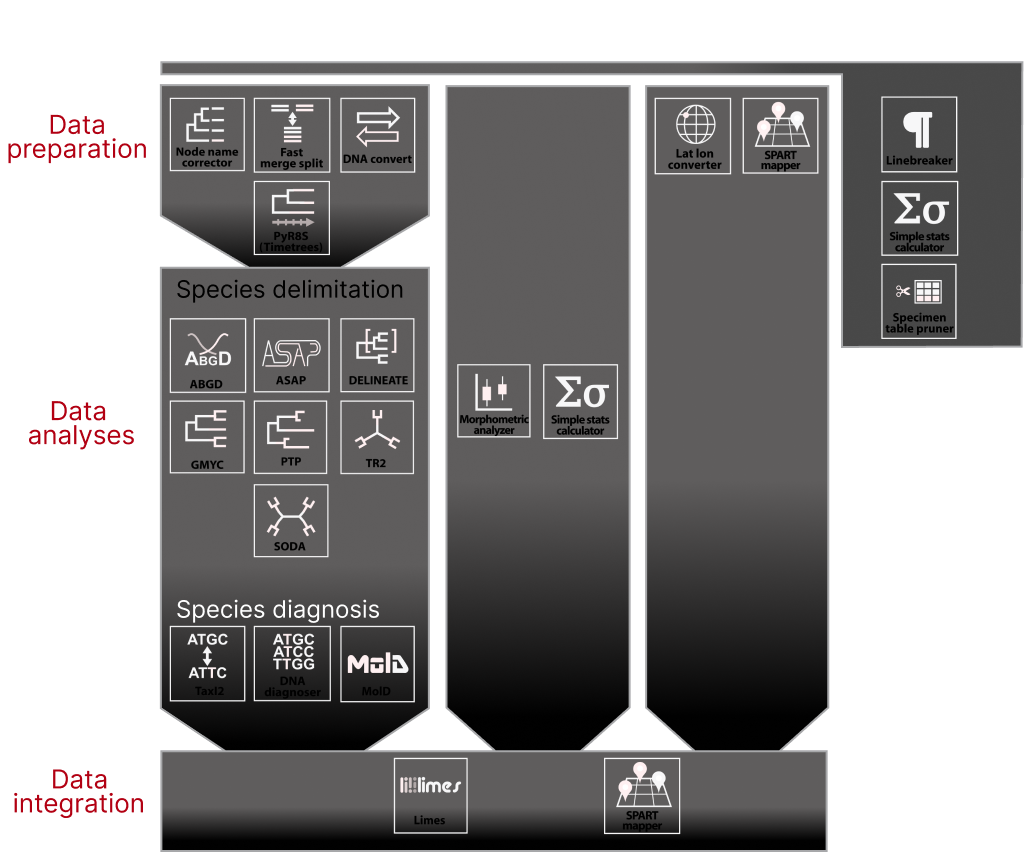
Conceived in 2021 in the framework of the Taxon-Omics priority program of the German Science Foundation, iTaxoTools is a bioinformatic toolkit targeted at facilitating alpha-taxonomic work, i.e., delimiting, diagnosing and describing species. Tools are largely built on open-source Python code with an emphasis on user-friendliness, i.e., intuitive graphical user interfaces, standalone executables for different computer platforms, as well as webservers. iTaxoTools is an ongoing project. The majority of tools will be continuously improved and new tools added, with an aim for more integrated species delimitation/description pipelines in the future. Check our “News” section below for updates..
Disclaimer: The programs included in iTaxoTools are free software. All code specifically programmed for iTaxoTools can be redistributed and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version. This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details. All tools not specifically programmed for iTaxoTools but constituting a modification or extension of the original code are also free software and most of them licensed under GNU v3, but partly, other licences apply. Please check the original GitHub pages for licensing of the original code of PTP. GMYC, tr2, DELINEATE, ABGD, ASAP, LIMES 2.0 and MolD.
Privacy statement: No personal data is collected, stored or processed on this website. No kind of user tracking is in use. We only use personal data provided to us (via email, phone, etc.) for the specified purpose that it was transmitted to us. Links to external websites are provided as a convenience and for informational purposes only; they do not constitute an endorsement or an approval of their content. We bear no responsibility for the accuracy, legality or content of the external site or for that of subsequent links.
.
.
News
28 February 2024
New version of Concatenator: Now includes Clipkit and a Python implementation of GBLOCKS to remove gappy stretches of sequences; available from the "Download" page.
30 January 2024
Publication of tutorial videos for ASAP, ABGD, MolD, and LIMES; videos on other tools will follow. See "Links" page for the links to the videos on Youtube.
23 January 2024
Preliminary launch of new tool "Hapsolutely" for haplotype phasing, reconstruction of haplotype genealogies and fields of recombination; now available from the "Download" page.
01 June 2023
Launch of the new, redesigned iTaxoTools website.
09 February 2023
Launch of a preliminary webserver using the Galaxy platform. Four tools (ASAP, MolD, DNAconvert, latlonconverter) have also been made available via the public toolshed of Galaxy. You can access the preliminary webserver here or from the link above.
02 February 2023
Launch of various revised tools: (1) new version of ASAP (now also as Mac executable), (2) MolD 1.4 with completely revised GUI (also for Mac), (3) New version of TaxI2 (version 1.0) with completely revised and much more user-friendly GUI as well as improved backend, (4) new version of DNAconvert with a few minor improvements.
01 December 2022
New tools made available: Concatenator, MAFFTpy, FastTreePy
23 July 2021
Publication of iTaxoTools paper in Megataxa
07 June 2021
Launch of iTaxotools.org
29 March 2021
Preprint of iTaxotools in BioRxiv
23 May 1707
Birth of Carl Linnaeus, historically presented as the "father of modern taxonomy", and to whom the iTaxotools logo (itself inspired by the portrait made by A. Roslin) pays homage
.
.
FAQ
One of the tools is not working on my computer, how should I proceed ?
Compiling executables that would work on all platforms (e.g., Windows 7, 8, 10 Home, 10 Professional, 11) is a challenge, and we cannot exclude that there might be problems on some platforms. If so, consider using the Python code itself - you will have to install Python and all necessary packages, and then can start the program directly from Python (including the GUI). This will usually be the most stable option and allows to change some settings (e.g., alignment parameters in DNAdiagnoser and TaxI2) that at present cannot be modified from the executables. Most tools should work reliably on Windows 10 but there might still be bugs on Windows 11.
I have discovered a real bug in one of the tools, what should I do ?
We recommend you sign up to GitHub, go to the repository of the respective tool, and create a "New Issue" there. The programmer of the respective tool will then correct the bug in the original code. In the next release, also the respective executable will be corrected.
Whom should I contact if I have a question ?
So far we do not yet have a dedicated Wiki, discussion group or support email. You can contact one of the lead scientists or developers, but it is recommended for now to submit your questions via GitHub Issues (see previous question).
Why aren't all tools available as webtools on the Galaxy server ?
Because some of the tools, such as TaxI2, are conceived as interactive tools which are difficult to implement in the Galaxy system, and others would be quite complex and time-consuming to use on Galaxy. We plan, however, to successively add more tools to the Galaxy system and Galaxy toolshed.
I produce home-made tools or I have innovative ideas of complementary approaches, can I contribute to the next version of iTaxoTools?
Yes, iTaxoTools is first and foremost a collaborative project designed for and by taxonomists, and jointly with various bioinformatics programmers. Any suggestion of new tools answering our philosophy is welcome!
Why, as a taxonomist, should I trust these automated delimitation tool whereas I can see that their results are sometimes very questionable?
The available species delimitation tools are only intended to explore the most likely species partitions within a given dataset and under specific assumptions. They allows taxonomists to infer species boundaries based on different detection criteria, such as barcode gap, coalescence, or monophyly, and will therefore very likely provide disparate results. The results inferred by a given method should therefore be considered with caution, and are to be regarded as an objective decision aid for partitioning individuals into initial species hypothesis that can then be subsequently tested in an integrative taxonomy pipeline, in combination with other datasets and methods.
Your initiative seems to be interesting, but actually it is useless for me (I am only working with morphological data !)
This first release of iTaxoTools is indeed mainly focused on molecular datasets, but new developments will allow to optimize morphological datasets and, if needed, to integrate them to molecular datasets.